NeuroShape combines a software interface and a digital anatomical atlas to create patient-tailored target maps of the thalamus. It is an extension to the popular medical image analysis software Slicer 3D.
What are the fields of application?
NeuroShape is a tool for academic research, and has not been approved by regulatory bodies. It can be used for a wide range of applications, such as providing additional visual guidance when creating neurosurgical target plans for MR-guided focused ultrasound neurosurgery or deep brain stimulation electrode implantation. Furthermore, the patient-tailored thalamus maps can serve as references to confirm the location of individual thalamic nuclei in research, such as in fMRI studies.
How is NeuroShape different from stereotactic atlases?
It relies on digitized 3D models from a multi-architectonic atlas of 7 thalami Krauth et al., 2010, previously developed by the ETH Zürich and University Hospital Zürich. Instead of using AC-PC or other reference points, the atlas is deformed to the patient's brain using MRI-visible structures as guidance, as described in a research paper by Jakab et al., 2012.
What are the benefits of using NeuroShape?
We hypothesize that with this approach, thalamus target maps are aligned better to the anatomy of the patient. This is very important when anatomy deviates from that of a stereotactic atlas: for example, in case of wide third ventricles, short AC-PC-distance, or any other observable anatomical deviation. Validation of the 3D SSM atlas is currently ongoing using high-resolution MRI and post mortem data.
Background
The planning of neurosurgical interventions requires great accuracy while localizing the target structures in the patient’s brain. In a typical setting, anatomical reference points are used on an MR image to transfer coordinates from a neuroanatomical atlas to the patient’s pre-operatively acquired images. This process can be improved in two ways: by extending the anatomical information that guides the transfer of coordinates, and by providing atlases that incorporate individual anatomical variability.
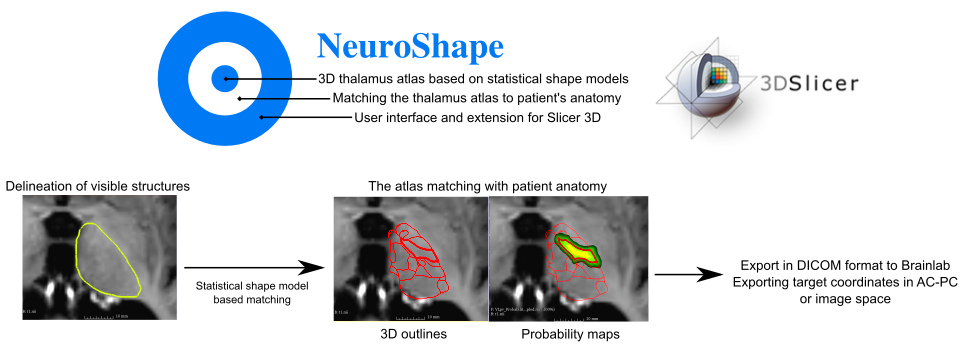
The neuroanatomical atlas used in the NeuroShape software is the multiarchitectonic analysis of 7 thalami as described in previous works (Krauth et al., 2010 and Morel, 1997), and is based on a more recent implementation of this atlas as a statistical shape model. The concept of the NeuroShape software is to use known correspondences between predictor and target structures from a statistical shape model atlas to provide target models for neurosurgical targeting that match the patient’s anatomy. Predictors are structures that are visible on targeting images, such as the macroscopic thalamus outlines on the pre- or intra-operatively acquired MRI. Instead of reference landmarks NeuroShape uses volumetric, 3D meshes as predictors. Individual thalamic nuclei, such as the ventrolateral posteroventral (VLpv – or Vim) nucleus, are often invisible on MRI, however, the geometry of these can be predicted using the correspondences to larger structures stored in the SSM atlas. Therefore NeuroShape can predict the geometry of the Vim nucleus of any patient given the thalamus outlines are manually delineated on the targeting MRI.
NeuroShape was realized as an academic collaboration between the Center for MR-Research, University Children's Hospital Zürich and the Computer Vision Laboratory, ETH Zürich.
The project “NeuroShape: a neurosurgical planning interface using statistical shape model of the thalamus” was supported by the Hasler Foundation and by the Institute of Physics and Engineering in Medicine (IPEM) Innovation Grant, awarded to PD Dr. Ruth Tuura.

If you use NeuroShape in your research, please cite the following references:
Krauth A, Blanc R, Poveda A, Jeanmonod D, Morel A, Székely G. A mean three-dimensional atlas of the human thalamus: generation from multiple histological data. Neuroimage. 2010 Feb 1;49(3):2053-62.
Jakab A, Blanc R, Berényi EL, Székely G. Generation of individualized thalamus target maps by using statistical shape models and thalamocortical tractography. AJNR Am J Neuroradiol. 2012 Dec;33(11):2110-6.




